The proton motive force occurs when the cell membrane becomes energized due to electron transport reactions by the electron carriers embedded in it. Basically, this causes the cell to act like a tiny battery. Its energy can either be used right away to do work, like power flagella, or be stored for later in ATP.
ATP synthesis is linked to the proton motive force through oxidative phosphorylation, where a phosphate group is added to ADP.
Although several steps are involved in creating an energized cell membrane, there’s one simple concept behind this phenomenon: the separation of positive protons (H+) on the outside of the membrane and negative hydroxide ions (OH–) on the inside of the membrane.
The fact that they’re charged makes it impossible for these ions to cross the membrane on their own. Trapping the ions on either side of the membrane creates two things, which together make the proton motive force: a pH and a charge difference. A difference in charge on the inside and the outside of a cell is called an electrochemical potential and is a huge source of energy.
Imagine you’re standing on the roof of a tall building, holding an orange. If you let the orange drop over the side of the building, by the time it reaches the ground it will have gained so much speed that it will hit the ground with great force and smash. Because of the large difference in height from where it was dropped and where it landed, there’s a great amount of energy.
It’s sort of the same thing with electrochemical potential, where the great difference in charge creates a lot of potential energy.
How this happens is that during electron transport, H+ are pushed to the outside of the membrane. The H+ come from both NADH and the dissociation of H2O into H+ and OH–. As you’ll see in a minute, it’s a bit more complicated, but the overall result is accumulation of a positive charge outside the cell and a negative charge within.
In the example, the proton motive force is created by a series of complexes within the cell membrane. These complexes are made up of the e– carriers mentioned earlier, the exact combination and number of which differ between organisms.
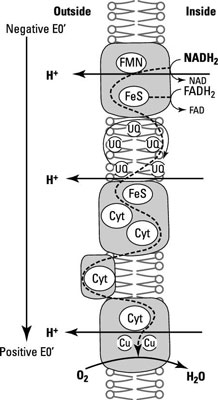
The proton motive force has two possible beginnings:
Complex I: One way the proton motive force begins is with the donation of H+ from NADH to flavin mononucleotide (FMN) to make FMNH2. 4H+ move to the outside of the cell when FMNH2 donates 2e– to the Fe/S proteins in complex I.
Complex II: The other way the proton motive force begins is through complex II, where FADH feeds e– and H+ from the oxidation of succinate, a product of the citric acid cycle, to the quinones. Complex II is less efficient than complex I.
Once electrons enter the cycle through Complex I or II they move on to Complex III and eventually IV:
Complex III: Quinones are reduced in the Q-cycle (a series of oxidation and reduction reactions of the coenzyme Q that result in the release of additional H+ to the outside of the membrane). Then electrons are passed one at a time from the Q-cycle to complex III, which contains the heme-containing proteins (specifically, cytochrome bc1) and an FeS protein.
Complex IV: Cytochrome bc1 transfers e– to cytochrome c, which then passes them to cytochromes a and a3 in complex IV. This is the end of the line where O2 is reduced to H2O.
At almost every step, H+ are pumped to the outer surface of the membrane, increasing the strength of the proton motive force.
Electron transport chains differ among organisms, but they always have three things in common:
Electron carriers arranged in order of increasing reducing power
Alternating H+ + e– and e–-only carriers
Generation of a proton motive force
The production of adenosine triphosphate (ATP) from aerobic respiration is called oxidative phosphorylation, and it’s carried out by a complex of proteins called ATP synthase.
This complex is made up of two subunits, F1 and F0, each of which is actually a rotary motor. Protons (H+), driven back through the membrane from the outside by the proton motive force, go through F0 and place pressure (or torque) on F1.
A molecule of adenosine diphosphate (ADP, with only two phosphate groups) along with a free phosphate group (Pi) bind to F1, and when the torque is released, energy is free to power the formation of ATP through the bonding of ADP and Pi. Like many enzymes, ATP synthase is reversible so that it can contribute to the proton motive force instead of weakening it.
So, even organisms that don’t use oxidative phosphorylation, like anaerobic fermenters, have ATP synthases so that they can still create a proton motive force to drive things like flagellar movement or ion transport.





